EnzFitter - 生物酶动力学曲线拟合软件
注:特此通知,BIOSOFT将于2022年12月31日停止交易。
电子邮件地址info@biosoft.com将在该日期后的2年内继续有效,以便向软件的注册用户提供程序激活代码和少量技术支持。
EnzFitter是一款通用的曲线拟合软件,适用于分析酶动力学实验。内置模块有或没有底物抑制/非竞争以及混合抑制的米式方程等模块,并可以自定义添加模块,适用于酶动力学实验分析,例如:通过对单个或两个基层数率的置信界限得到开始的比率和参数值。
EnzFitter内置的功能,可以批处理模式运行,并且OLE自动化。可以根据拟合的标准曲线计算未知数。可以创建附加到文件,数据或曲线拟合的文本注释。
Biosoft EnzFitter主要:
1. EnzFitter是一种通用的曲线拟合软件包,具有设计用于适用于酶动力学实验分子的自定义功能。例如,初始速率和参数值可以通过他们对于单基质速率数据和双基质速率数据的置信限来获得。内置模型具有或不具有底物抑制作用的Michaelis-Menten,竞争性,无竞争性和混合抑制作用,三元复合物或有序bi-bi系统以及有和无底物抑制的乒乓球。您可以使用传统的代数语法添加模型。
2. 数据导入格式EnzFitter for DOS文件以及ASCII分隔符,dBase,FoxPro,Excel,Quattro Pro,Lotus123,Paradox和Symphony。文件和数据集大小可用内存的限制,并且可以为文件创建多个数据集。有数据编辑功能,:插入/编辑/删除点,复制和移动数据块和编辑列标题。照顾缺少数据和复制品。可以显示和导出数据的描述性统计工具。工作表使可自定义的(例如字体,颜色,边框,行高和列宽),当打印数据时,可以给自定义页眉或页脚。您可以随时插入新的列或行。提供了数据转换,或者您可以创建自己的转换方程。您可以为经常使用的数据格式创建模板,并结合图模板实现常规检测的自动化。
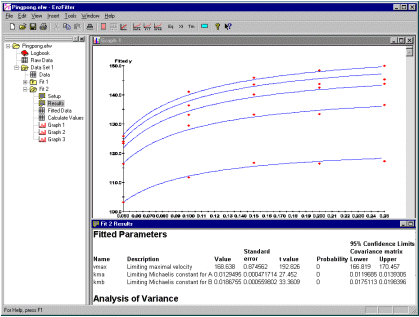
3. 可以使用Marquardt或Simplex算法对数据集执行多重曲线拟合。可以对数据进行稳健,统计,比例或显式值加权。遗传算法可以生成出色的初始参数估计值,但参数可以被限制在一定范围内,或者在时固定。结果以表格和图形形式呈现,并可以绘制原始数据和拟合数据的推行,并且为了区分数据集,您可以选择符号,半连续线和添加标签。可以在主图上插入额外数据集的附属图,相同数据的残差和变换/衍生图。
4. EnzFitter可以设置为以批处理模式运行(自动执行多个分析,用户干预),并且还OLE自动化。对于更的化验用途,您可以根据拟合的标准曲线计算未知数。您可以创建附加到文件,数据集或曲线拟合的文本注释。还有自动记录日志记录对给定文件所做的更改。
EnzFitter 功能
-
EnzFitter 提供的方程
-
Michaelis-Menten
-
竞争性抑制
-
非竞争性抑制作用
-
没有竞争力的抑制
-
混合抑制
-
三元络合机理(有序 Bi-Bi)
-
底物抑制
-
Ping-Pong Bi-Bi 系统(无抑制)
-
Ping-Pong Bi-Bi系统(A抑制)
-
Ping-Pong Bi-Bi系统(B抑制)
-
Ping-Pong Bi-Bi 系统(A 和 B 抑制)
-
线性回归
-
一号订单率
-
pka测定
-
配体结合(1 个站点)
-
配体结合(1 个带有 NSB 的站点)
-
配体结合(2 个站点)
-
配体结合(2 个带有 NSB 的站点)
-
单指数衰减
-
双指数衰减
-
三重指数衰减
-
变构动力学(Hill方程)
-
不对称 Sigmoid
-
双线性
-
立方体
-
立方(参数)
-
三次(根)
-
指数 Sigmoid
-
Gompertz 增长
-
日志物流增长
-
对数Logistic增长与X偏移
-
逻辑 Sigmoid
-
Morgan-Mercer-Florin 增长
-
具有 X 和 Y 偏移量的幂次定律
-
二次方
-
二次参数
-
二次根
-
理查兹成长
-
统计 Sigmoid
-
三相线性
-
两相线性
在 Enzfitter 中自动化的一些转换/导数图
-
Residuals
-
Scatchard
-
Eadie
-
Lineweaver-Burk
-
线性化 pKa
【英文介绍】
Enzyme Kinetics and Curve Fitting
EnzFitter is a generic curve-fitting package which has custom features designed to make it especially suitable for analysis of enzyme kinetics experiments. For example, initial rate and parameter values can be obtained with their confidence limits for single and twin substrate rate data. Built-in models include Michaelis-Menten with or without substrate inhibition, competitive, uncompetitive and mixed inhibition, ternary complex or ordered bi-bi systems and ping-pong with and without inhibition by substrates. You can easily add other models in conventional algebraic syntax.
Data import formats include EnzFitter for DOS files as well as ASCII delimited, dBase, FoxPro, Excel, Quattro Pro, Lotus 123, Paradox and Symphony. File and dataset sizes are limited only by available memory and multiple data sets can be created for each file. There are advanced data editing facilities including: insert/edit/remove points, copy and move blocks of data and edit column titles. Missing data and replicates are catered for. Descriptive statistics of the data can be displayed and exported. The worksheet is customizable (e.g. font, colors, borders, row height and column width) and when data are printed they can be given custom headers or footers. You can insert new columns or rows at any time. Many data transforms are supplied or you can create your own equations for transforms. You can create templates for data formats you use frequently which, combined with graph templates virtually automate routine assays.

Multiple curve fits can be performed for any data set using either the Marquardt or Simplex algorithms. Optionally, data can be weighted robustly, statistically, proportionally or with explicit values. A genetic algorithm generates excellent initial parameter estimates but parameters can be constrained to a range of values or fixed when necessary. The results are presented in tabular and graphic form and can be saved to disk, printed or exported to other programs. Export formats include, for graphs, Bitmaps, Metafiles, JPG or TIF and, for numeric results, ASCII delimited and Excel. Graphs of raw and fitted data can be plotted and, to distinguish datasets, you can select a variety of symbols, semi-continuous lines and also add labels. Subsidiary graphs of extra sets of data, residuals and transformed /derivative plots of the same data can be inset on the main graph.
EnzFitter can be set up to run in batch mode (performing several analyses automatically, without user intervention) and it also supports OLE Automation. For more general assay use, you can calculate unknowns from fitted standard curves. You can create text Notes that are attached to any file, dataset or curve fit. There is also an automatic Logbook which records all changes made to a given file.
EnzFitter Features
-
Equations provided with EnzFitter
-
Michaelis-Menten
-
Competitve Inhibition
-
Non-Competitive Inhibition
-
Uncompetitive Inhibition
-
Mixed Inhibition
-
Ternary Complex Mechanism(Ordered Bi-Bi)
-
Substrate Inhibition
-
Ping-Pong Bi-Bi System(No Inhibition)
-
Ping-Pong Bi-Bi System(Inhibition by A)
-
Ping-Pong Bi-Bi System(Inhibition by B)
-
Ping-Pong Bi-Bi System(Inhibition by A and B)
-
Linear Regression
-
1st Order Rate
-
pka Determination
-
Ligand Binding(1 Site)
-
Ligand Binding(1 Site with NSB)
-
Ligand Binding(2 Sites)
-
Ligand Binging(2 Sites with NSB)
-
Single Exponential Decay
-
Double Exponential Decay
-
Triple Exponential Decay
-
Allosteric Kinetics(Hill Equation)
-
Asymmetric Sigmoid
-
Bilinear
-
Cubic
-
Cubic(Parametric)
-
Cubic(Roots)
-
Exponential Sigmoid
-
Gompertz Growth
-
Log Logistic Growth
-
Log Logistic Growtg with X Offset
-
Logisitic Sigmoid
-
Morgan-Mercer-Florin Growth
-
Power Law with X and Y Offaets
-
Quadratic
-
Quadratic Parametric
-
Quadratic Roots
-
Richards Growth
-
Statistical Sigmoid
-
Three-Phase Linear
-
Two-Phase Linear
Some of the transformed/derivative plots automated in Enzfitter
-
Residuals
-
Scatchard
-
Eadie
-
Lineweaver-Burk
-
Linearize pKa
- 2026-02-09
- 2026-01-20
- 2026-01-16
- 2026-01-12
- 2026-01-12
- 2026-01-09
- 2026-02-05
- 2026-02-05
- 2026-01-28
- 2026-01-26
- 2026-01-26
- 2026-01-16